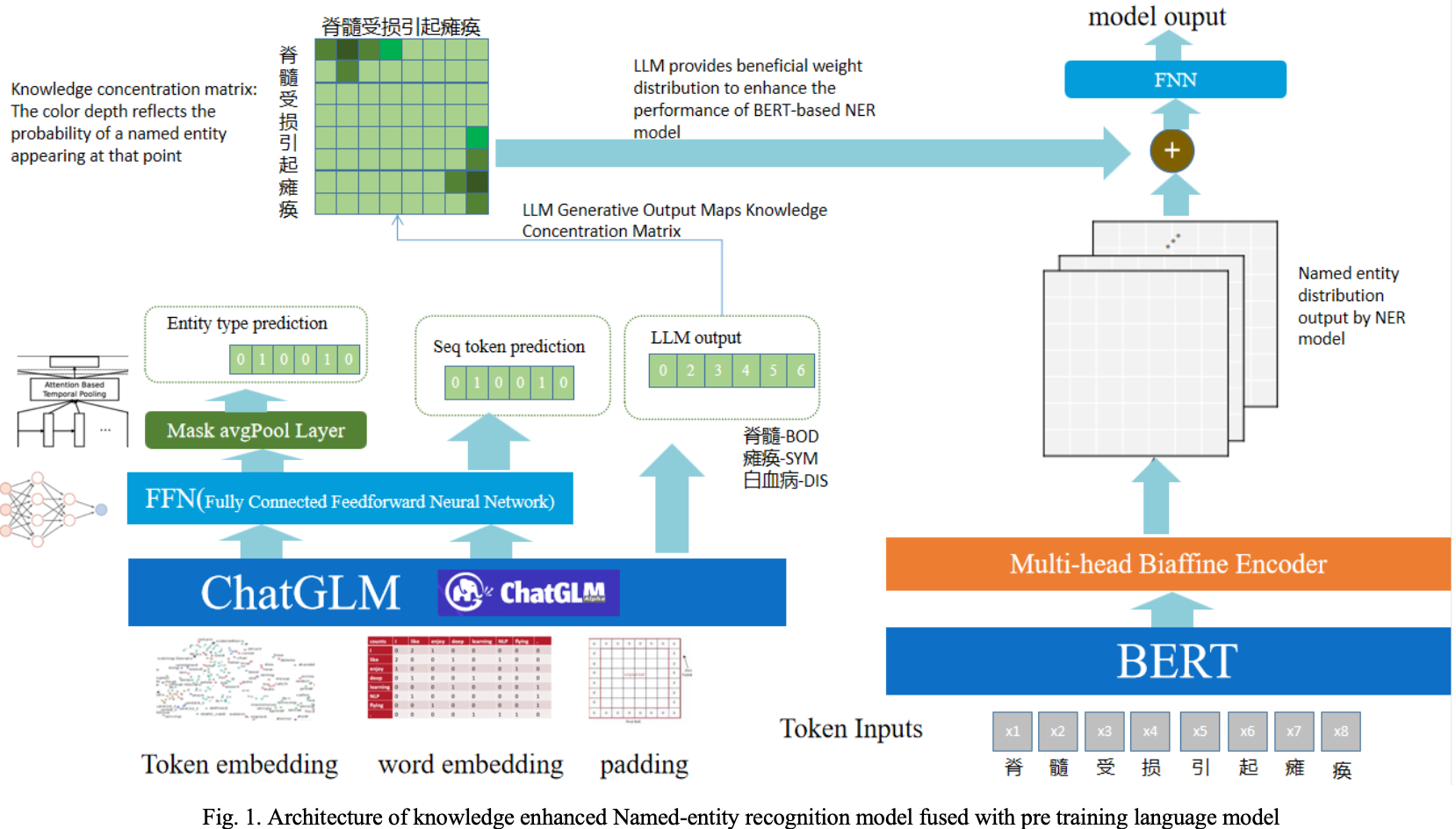
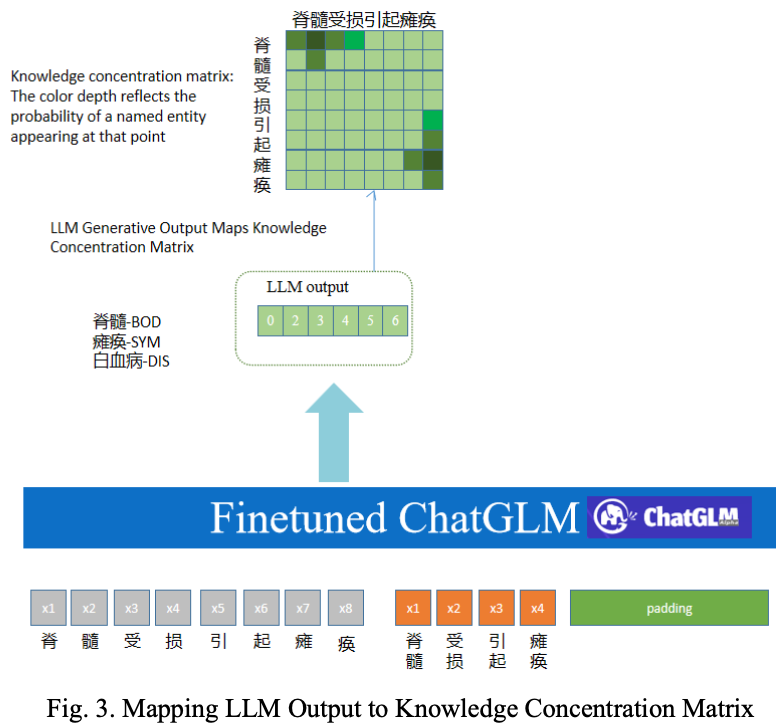
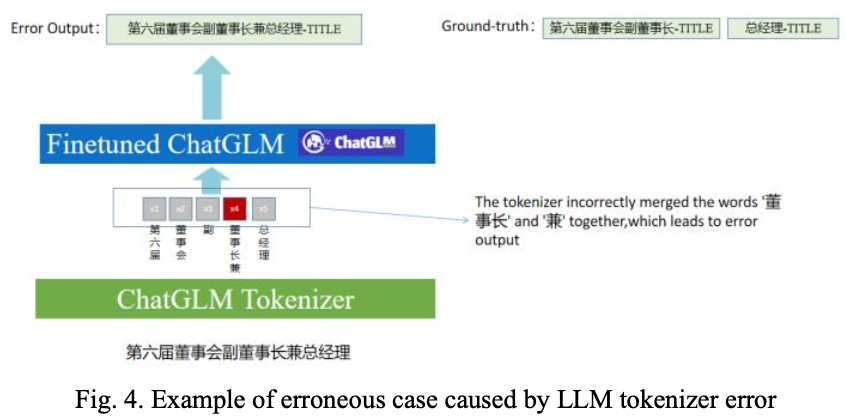
A Knowledge-enhanced Medical Named Entity Recognition Method that Integrates Pre-trained Language Models
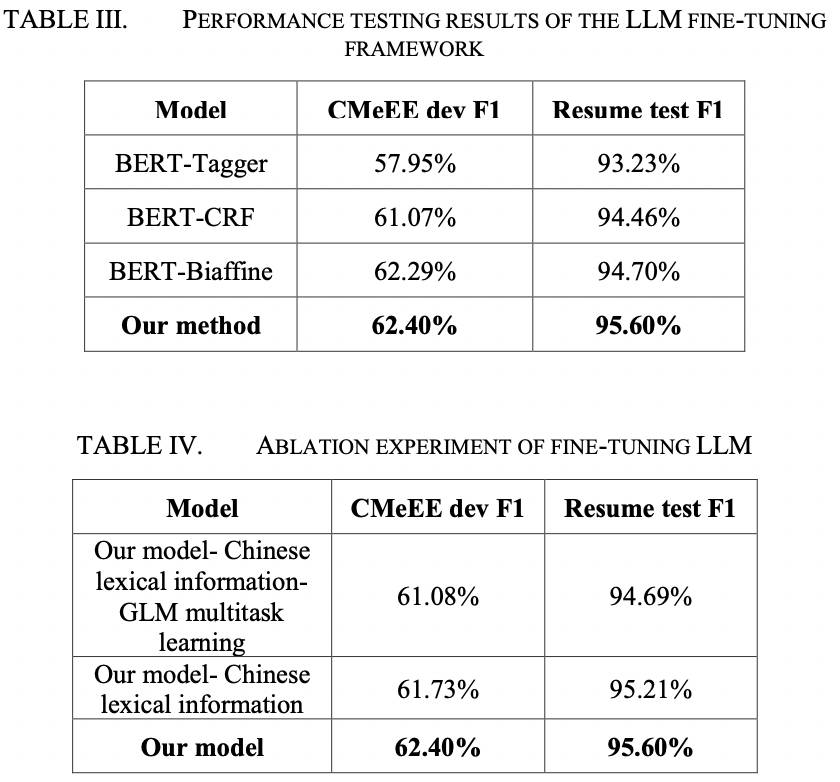
Medical Named Entity Recognition (NER) is a critical task in medical text processing that involves identifying and classifying specific entities within medical documents. Accurate and automated recognition of these entities is essential for various downstream applications, including clinical decision support systems, electronic health records and biomedical literature analysis. Medical documents exhibit high variability in terms of language usage, abbreviations, synonyms, misspellings, and typographical errors, so the precise extraction of named entities is challenging. Although large language models (LLMs) have shown good performance in medical knowledge extraction tasks in few-shot settings, their performance is difficult to fully leverage in supervised medical named entity recognition (NER) tasks. This is because NER is a sequence labeling task, while LLMs are more suitable for tasks such as text generation. Furthermore, the structured output of NER tasks leads to a performance loss when LLMs convert it into generative text. Therefore, it is a challenging problem to utilize LLMs to improve the accuracy of medical named entity recognition tasks. On this paper, we propose a method that integrates LLM knowledge to enhance the performance of medical NER models. Firstly, we improve the structure of the LLM model to make it more adaptable to named entity sequence labeling and preserve the structural characteristics of NER tasks. Secondly, we adopt the LoRA fine-tuning method and incorporate Chinese vocabulary information into the model training, making the training process more efficient. Finally, to fully utilize the fine-tuned LLM to enhance the medical NER model, we convert the output of the LLM into a knowledge concentration matrix and inject it into the NER model. We have verify the effectiveness of our new method on the CMeEE dataset. The results demonstrate that our method can efficiently fine-tune the LLM and improve its performance in medical named entity recognition tasks. Moreover, our method can also leverage the prior knowledge of the LLM to enhance the BERT-based medical NER model. In addition, our method demonstrates good generalization and can tackle entity recognition tasks in other domains. We validated the superiority of our approach on the resume-zh dataset.